Product References
Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity.
Nature
Meng B,Abdullahi A,Ferreira IATM,Goonawardane N,Saito A,Kimura I,Yamasoba D,Gerber PP,Fatihi S,Rathore S,Zepeda SK,Papa G,Kemp SA,Ikeda T,Toyoda M,Tan TS,Kuramochi J,Mitsunaga S,Ueno T,Shirakawa K,Takaori-Kondo A,Brevini T,Mallery DL,Charles OJ,Bowen JE,Joshi A,Walls AC,Jackson L,Martin D,Smith KGC,Bradley J,Briggs JAG,Choi J,Madissoon E,Meyer KB,Mlcochova P,Ceron-Gutierrez L,Doffinger R,Teichmann SA,Fisher AJ,Pizzuto MS,de Marco A,Corti D,Hosmillo M,Lee JH,James LC,Thukral L,Veesler D,Sigal A,Sampaziotis F,Goodfellow IG,Matheson NJ,Sato K,Gupta RK
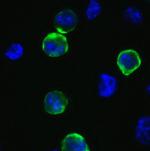
PA5-112048 was used in Flow Cytometry to show that the spike protein of Omicron has a higher affinity for ACE2 compared with Delta, and a marked change in its antigenicity increases Omicron's evasion of therapeutic monoclonal and vaccine-elicited polyclonal neutralizing antibodies after two doses.
Tue Mar 01 00:00:00 EST 2022
Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity.
Nature
Meng B,Abdullahi A,Ferreira IATM,Goonawardane N,Saito A,Kimura I,Yamasoba D,Gerber PP,Fatihi S,Rathore S,Zepeda SK,Papa G,Kemp SA,Ikeda T,Toyoda M,Tan TS,Kuramochi J,Mitsunaga S,Ueno T,Shirakawa K,Takaori-Kondo A,Brevini T,Mallery DL,Charles OJ,Bowen JE,Joshi A,Walls AC,Jackson L,Martin D,Smith KGC,Bradley J,Briggs JAG,Choi J,Madissoon E,Meyer KB,Mlcochova P,Ceron-Gutierrez L,Doffinger R,Teichmann SA,Fisher AJ,Pizzuto MS,de Marco A,Corti D,Hosmillo M,Lee JH,James LC,Thukral L,Veesler D,Sigal A,Sampaziotis F,Goodfellow IG,Matheson NJ,Sato K,Gupta RK
PA5-112048 was used in Flow Cytometry to show that the spike protein of Omicron has a higher affinity for ACE2 compared with Delta, and a marked change in its antigenicity increases Omicron's evasion of therapeutic monoclonal and vaccine-elicited polyclonal neutralizing antibodies after two doses.
Tue Mar 01 00:00:00 EST 2022
The SARS-CoV-2 spike S375F mutation characterizes the Omicron BA.1 variant.
iScience
Kimura I,Yamasoba D,Nasser H,Zahradnik J,Kosugi Y,Wu J,Nagata K,Uriu K,Tanaka YL,Ito J,Shimizu R,Tan TS,Butlertanaka EP,Asakura H,Sadamasu K,Yoshimura K,Ueno T,Takaori-Kondo A,Schreiber G,Toyoda M,Shirakawa K,Irie T,Saito A,Nakagawa S,Ikeda T,Sato K
PA5-112048 was used in Flow Cytometry to elucidate that the F375 residue forms an interprotomer pi-pi interaction with the H505 residue of another protomer in the spike trimer, conferring the attenuated cleavage efficiency and fusogenicity of Omicron spike.
Thu Dec 22 00:00:00 EST 2022
Multiple expansions of globally uncommon SARS-CoV-2 lineages in Nigeria.
Nature communications
Ozer EA,Simons LM,Adewumi OM,Fowotade AA,Omoruyi EC,Adeniji JA,Olayinka OA,Dean TJ,Zayas J,Bhimalli PP,Ash MK,Maiga AI,Somboro AM,Maiga M,Godzik A,Schneider JR,Mamede JI,Taiwo BO,Hultquist JF,Lorenzo-Redondo R
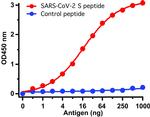
PA5-112048 was used in ELISA to demonstrate a distinct distribution of SARS-CoV-2 lineages in Nigeria, and emphasise the need for improved genomic surveillance worldwide.
Thu Feb 03 00:00:00 EST 2022
Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell
Kimura I,Yamasoba D,Tamura T,Nao N,Suzuki T,Oda Y,Mitoma S,Ito J,Nasser H,Zahradnik J,Uriu K,Fujita S,Kosugi Y,Wang L,Tsuda M,Kishimoto M,Ito H,Suzuki R,Shimizu R,Begum MM,Yoshimatsu K,Kimura KT,Sasaki J,Sasaki-Tabata K,Yamamoto Y,Nagamoto T,Kanamune J,Kobiyama K,Asakura H,Nagashima M,Sadamasu K,Yoshimura K,Shirakawa K,Takaori-Kondo A,Kuramochi J,Schreiber G,Ishii KJ,Hashiguchi T,Ikeda T,Saito A,Fukuhara T,Tanaka S,Matsuno K,Sato K
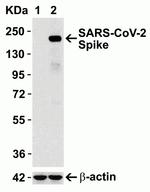
PA5-112048 was used in ELISA, Western Blot, Neutralization to suggest that the risk of BA.2 subvariants, particularly BA.4/5, to global health is greater than that of original BA.2.
Thu Oct 13 00:00:00 EDT 2022
Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell
Kimura I,Yamasoba D,Tamura T,Nao N,Suzuki T,Oda Y,Mitoma S,Ito J,Nasser H,Zahradnik J,Uriu K,Fujita S,Kosugi Y,Wang L,Tsuda M,Kishimoto M,Ito H,Suzuki R,Shimizu R,Begum MM,Yoshimatsu K,Kimura KT,Sasaki J,Sasaki-Tabata K,Yamamoto Y,Nagamoto T,Kanamune J,Kobiyama K,Asakura H,Nagashima M,Sadamasu K,Yoshimura K,Shirakawa K,Takaori-Kondo A,Kuramochi J,Schreiber G,Ishii KJ,Hashiguchi T,Ikeda T,Saito A,Fukuhara T,Tanaka S,Matsuno K,Sato K
PA5-112048 was used in ELISA, Western Blot, Neutralization to suggest that the risk of BA.2 subvariants, particularly BA.4/5, to global health is greater than that of original BA.2.
Thu Oct 13 00:00:00 EDT 2022
Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell
Kimura I,Yamasoba D,Tamura T,Nao N,Suzuki T,Oda Y,Mitoma S,Ito J,Nasser H,Zahradnik J,Uriu K,Fujita S,Kosugi Y,Wang L,Tsuda M,Kishimoto M,Ito H,Suzuki R,Shimizu R,Begum MM,Yoshimatsu K,Kimura KT,Sasaki J,Sasaki-Tabata K,Yamamoto Y,Nagamoto T,Kanamune J,Kobiyama K,Asakura H,Nagashima M,Sadamasu K,Yoshimura K,Shirakawa K,Takaori-Kondo A,Kuramochi J,Schreiber G,Ishii KJ,Hashiguchi T,Ikeda T,Saito A,Fukuhara T,Tanaka S,Matsuno K,Sato K
PA5-112048 was used in ELISA, Western Blot, Neutralization to suggest that the risk of BA.2 subvariants, particularly BA.4/5, to global health is greater than that of original BA.2.
Thu Oct 13 00:00:00 EDT 2022
Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell
Kimura I,Yamasoba D,Tamura T,Nao N,Suzuki T,Oda Y,Mitoma S,Ito J,Nasser H,Zahradnik J,Uriu K,Fujita S,Kosugi Y,Wang L,Tsuda M,Kishimoto M,Ito H,Suzuki R,Shimizu R,Begum MM,Yoshimatsu K,Kimura KT,Sasaki J,Sasaki-Tabata K,Yamamoto Y,Nagamoto T,Kanamune J,Kobiyama K,Asakura H,Nagashima M,Sadamasu K,Yoshimura K,Shirakawa K,Takaori-Kondo A,Kuramochi J,Schreiber G,Ishii KJ,Hashiguchi T,Ikeda T,Saito A,Fukuhara T,Tanaka S,Matsuno K,Sato K
PA5-112048 was used in ELISA, Western Blot, Neutralization to suggest that the risk of BA.2 subvariants, particularly BA.4/5, to global health is greater than that of original BA.2.
Thu Oct 13 00:00:00 EDT 2022
Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell
Kimura I,Yamasoba D,Tamura T,Nao N,Suzuki T,Oda Y,Mitoma S,Ito J,Nasser H,Zahradnik J,Uriu K,Fujita S,Kosugi Y,Wang L,Tsuda M,Kishimoto M,Ito H,Suzuki R,Shimizu R,Begum MM,Yoshimatsu K,Kimura KT,Sasaki J,Sasaki-Tabata K,Yamamoto Y,Nagamoto T,Kanamune J,Kobiyama K,Asakura H,Nagashima M,Sadamasu K,Yoshimura K,Shirakawa K,Takaori-Kondo A,Kuramochi J,Schreiber G,Ishii KJ,Hashiguchi T,Ikeda T,Saito A,Fukuhara T,Tanaka S,Matsuno K,Sato K
PA5-112048 was used in ELISA, Western Blot, Neutralization to suggest that the risk of BA.2 subvariants, particularly BA.4/5, to global health is greater than that of original BA.2.
Thu Oct 13 00:00:00 EDT 2022
The SARS-CoV-2 Omicron BA.1 spike G446S mutation potentiates antiviral T-cell recognition.
Nature communications
Motozono C,Toyoda M,Tan TS,Hamana H,Goto Y,Aritsu Y,Miyashita Y,Oshiumi H,Nakamura K,Okada S,Udaka K,Kitamatsu M,Kishi H,Ueno T
PA5-112048 was used in Western Blotting to demonstrate that the G446S mutation in the Omicron BA.1 variant affects antigen processing/presentation and potentiates antiviral activity by vaccine-induced T cells, leading to enhanced T cell recognition towards emerging variants.
Wed Sep 21 00:00:00 EDT 2022
Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant.
Nature
Suzuki R,Yamasoba D,Kimura I,Wang L,Kishimoto M,Ito J,Morioka Y,Nao N,Nasser H,Uriu K,Kosugi Y,Tsuda M,Orba Y,Sasaki M,Shimizu R,Kawabata R,Yoshimatsu K,Asakura H,Nagashima M,Sadamasu K,Yoshimura K,Sawa H,Ikeda T,Irie T,Matsuno K,Tanaka S,Fukuhara T,Sato K
PA5-112048 was used in ELISA, Western Blot to reveal the virological characteristics of Omicron, including rapid growth in the human population, lower fusogenicity and attenuated pathogenicity.
Tue Mar 01 00:00:00 EST 2022
The SARS-CoV-2 Lambda variant exhibits enhanced infectivity and immune resistance.
Cell reports
Kimura I,Kosugi Y,Wu J,Zahradnik J,Yamasoba D,Butlertanaka EP,Tanaka YL,Uriu K,Liu Y,Morizako N,Shirakawa K,Kazuma Y,Nomura R,Horisawa Y,Tokunaga K,Ueno T,Takaori-Kondo A,Schreiber G,Arase H,Motozono C,Saito A,Nakagawa S,Sato K
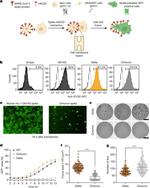
PA5-112048 was used in Flow Cytometry to reveal that the spike protein of the Lambda variant is more infectious than that of other variants due to the T76I and L452Q mutations.
Tue Jan 11 00:00:00 EST 2022
Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation.
Nature
Saito A,Irie T,Suzuki R,Maemura T,Nasser H,Uriu K,Kosugi Y,Shirakawa K,Sadamasu K,Kimura I,Ito J,Wu J,Iwatsuki-Horimoto K,Ito M,Yamayoshi S,Loeber S,Tsuda M,Wang L,Ozono S,Butlertanaka EP,Tanaka YL,Shimizu R,Shimizu K,Yoshimatsu K,Kawabata R,Sakaguchi T,Tokunaga K,Yoshida I,Asakura H,Nagashima M,Kazuma Y,Nomura R,Horisawa Y,Yoshimura K,Takaori-Kondo A,Imai M,Tanaka S,Nakagawa S,Ikeda T,Fukuhara T,Kawaoka Y,Sato K
PA5-112048 was used in ELISA, Western Blot to demonstrate that the P681R-bearing virus exhibits higher pathogenicity compared with its parental virus.
Tue Feb 01 00:00:00 EST 2022
Variations in cell-surface ACE2 levels alter direct binding of SARS-CoV-2 Spike protein and viral infectivity: Implications for measuring Spike protein interactions with animal ACE2 orthologs.
bioRxiv : the preprint server for biology
Kazemi S,López-Muñoz AD,Hollý J,Jin L,Yewdell JW,Dolan BP
PA5-112048 was used in Western Blot to describe a method to generate cells stably expressing equivalent levels of different ACE2 orthologs, the receptor for SARS-CoV-2, on the surface of a human cell line.
Fri Oct 22 00:00:00 EDT 2021