Article by Aparna Chandrasekaran
Introduction to PRMTs
Protein arginine methyltransferases (PRMTs) are ‘writers’ of arginine methylation in histone and non-histone proteins. PRMTs are divided into three types, based on whether they promote monomethylation (MMA), asymmetric (ADMA), or symmetric dimethylation (SDMA) of arginine. These three derivatives are present on a multitude of diverse protein classes in the cytoplasm, nucleus, and organelles of cells.
PRMTs transfer methyl groups from S-adenosylmethionine (AdoMet) to the guanidino nitrogen of arginine residue. When two methyl groups are located on one of the terminal nitrogen atoms of the guanidino group, it leads to the formation of the asymmetric dimethylarginine derivative (ADMA). When the methyl group is placed on each of the terminal guanidino nitrogen atoms, it produces the symmetric dimethylarginine derivative (SDMA), and when a single methyl group is found on the terminal nitrogen atom, it is known as the monomethylated Arginine (MMA).
In humans, nine PRMTs have been identified so far with the type I arginine methyltransferases consisting of PRMT1, PRMT2, PRMT3, PRMT4, PRMT6, and PRMT8, type II consisting of PRMT5, and PRMT9 that catalyze the formation of ADMA and SDMA respectively. The type III enzyme consists of PRMT7 that only catalyzes the formation of MMA.
Table 1 Invitrogen antibodies for studying PRMTs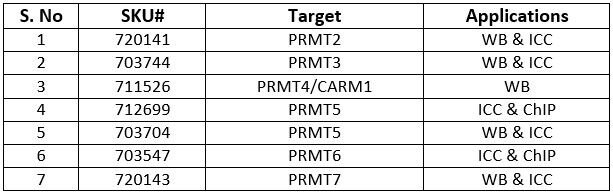
Role of PRMTs
The ability of PRMTs to catalyze arginine methylation results in changes in protein-protein interactions involved in biological processes such as cell signaling, cell differentiation, DNA repair, mRNA splicing, transcription, mRNA translation, and embryonic development.
The main substrates of PRMTs are histones leading to their foremost role in the epigenetic regulation of gene expression. They methylate key activating (histone H4R3me2a, H3R2me2s, H3R17me2a, H3R26me2a) and repressive histone marks (H3R2me2a, H3R8me2a, H3R8me2s, H4R3me2s).
The table below briefly mentions the methylation substrates and functions of each of the PRMTs.
Table 2 Function of PRMTs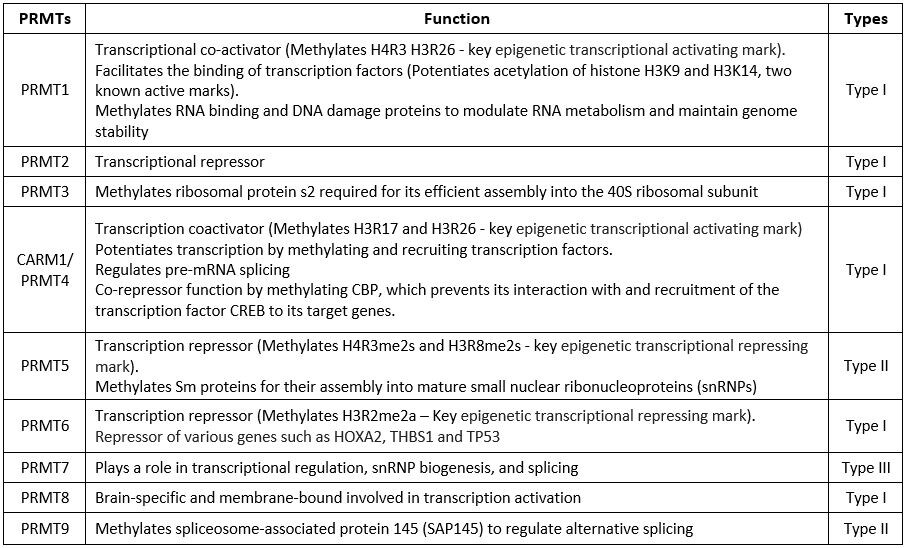
We offer specific antibodies against some of the key target PRMTs (Table 1) which have been validated by genetic modification strategies where reduction or abrogation of target expression was used to demonstrate antibody specificity.
Figure 1 Western blot was performed using A) Anti-PRMT2 Rabbit Polyclonal Antibody (Product # 720141), B) Anti-PRMT3 Recombinant Rabbit Monoclonal Antibody (Product # 703744) and C) Anti-PRMT5 Recombinant Rabbit Monoclonal Antibody (Product # 703704) and the band of interest corresponding to the protein was observed across the cell lines tested.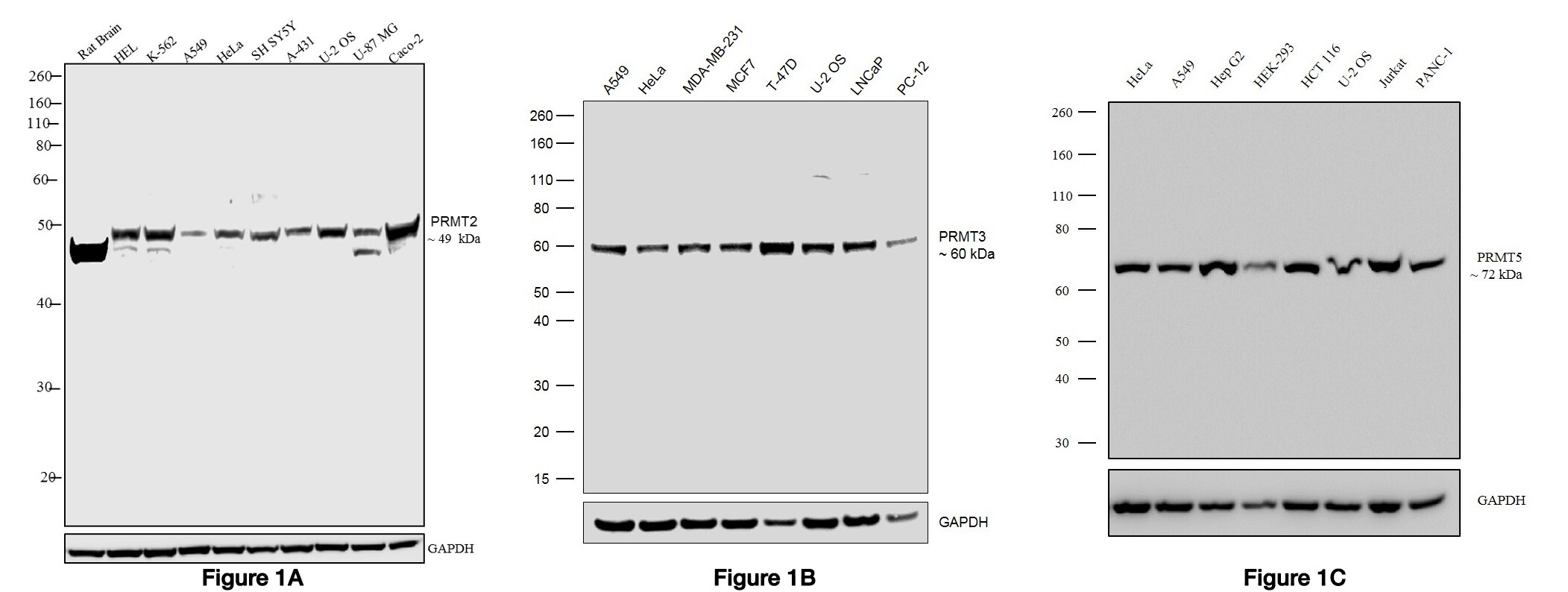
Figure 2 Knockdown of PRMT5 and PRMT3 was achieved by transfecting MCF-7 and A549 cells with PRMT specific siRNAs. Immunofluorescence analysis of PRMT5 (Figure 3 A) and PRMT3 (Figure 3 B) was performed on MCF-7 and A549 cells (untransfected, panel a-d), transfected with PRMT5 and PRMT3 specific siRNA (panel i-l) or non-specific scrambled siRNA (panels e-h) respectively. The cells were labeled with A) Anti-PRMT5 Recombinant Rabbit Polyclonal Antibody (Product # 712669) and B) Anti-PRMT3 Recombinant Rabbit Monoclonal Antibody (Product # 703744). Chemiluminescent detection was performed for both western blots using Thermo Scientific™ Pierce™ ECL Western Blotting Substrate (Cat. No. #32106) and the Invitrogen™ iBright™ Imaging System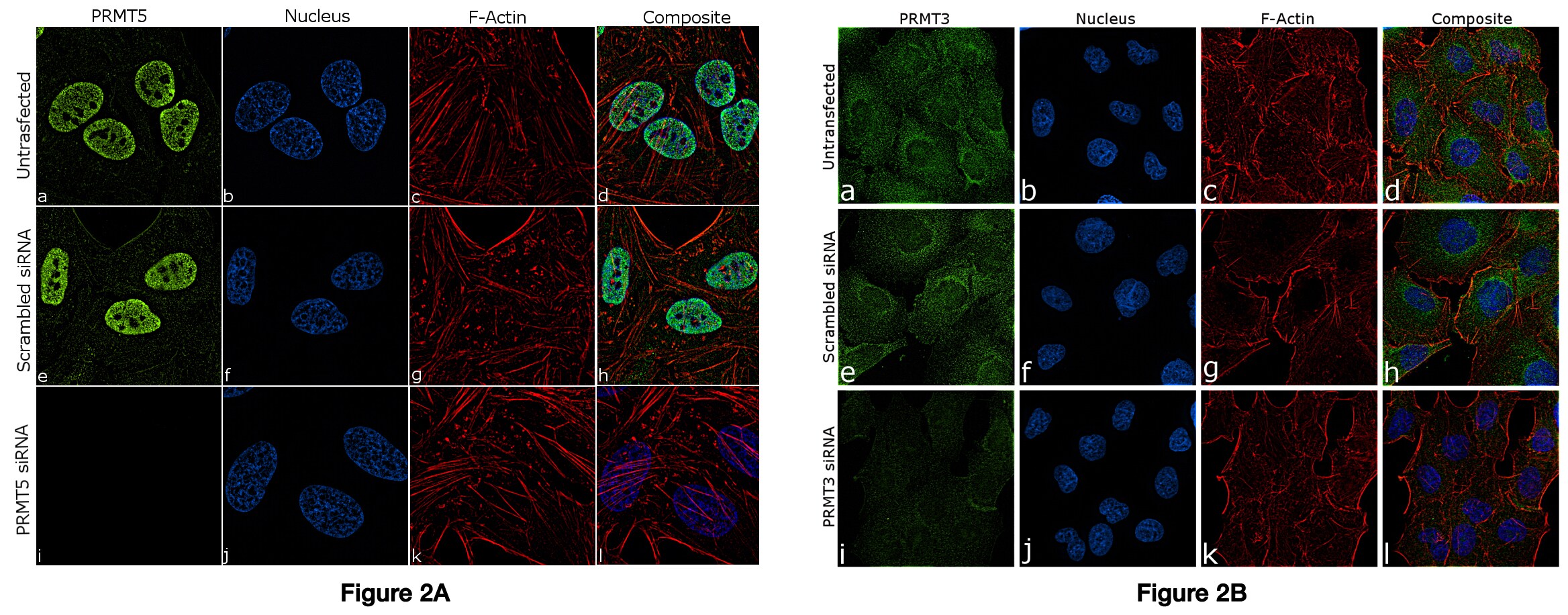
Currently, the method of choice for genomic mapping of PRMTs and their arginine methylated substrates is chromatin immunoprecipitation (ChIP) shown in Figure 3. It helps to understand the molecular mechanisms eliciting the PRMT-mediated transcriptional regulation.
Figure 3 Chromatin Immunoprecipitation (ChIP) was performed using A) Anti-PRMT5 Recombinant Rabbit Polyclonal Antibody (Product # 712669) using primers binding to active (ADIPOQ, BIRC3, RETN) and inactive (SAT2 satellite repeats loci. B) Anti-PRMT6 Recombinant Rabbit Monoclonal Antibody (Product # 703547) using primers binding to active (CDKN1A, HXA2) and inactive (SATA satellite repeats) loci. 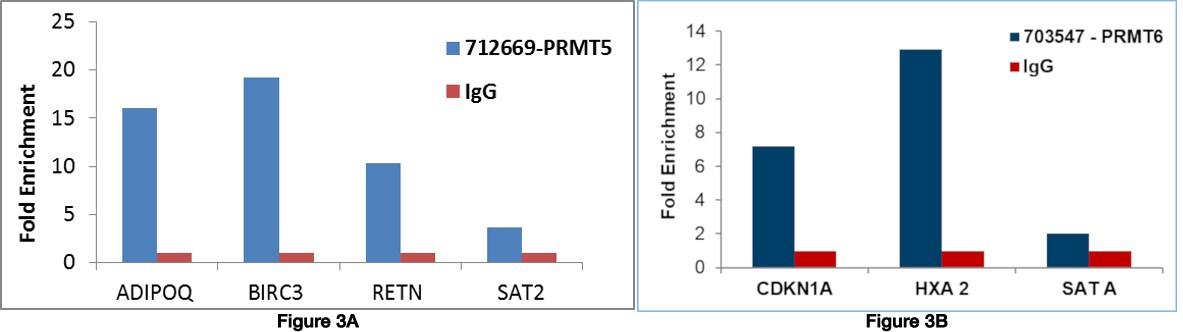
PRMTs and Cancer
Arginine methylation is an understudied modification that is increasingly linked with deregulation in cancer. Processes hijacked by cancer cell to ensure survival, include epigenetic-mediated gene expression, mRNA splicing, and the DNA damage response which are also regulated by PRMTs. Consequently, PRMTs, the writers of arginine methylation in clinical prospect have rapidly gained interest as novel drug targets.
Mutations in PRMT genes leading to cancer are rare, but their expression is often upregulated in cancer and associated with poor patient prognosis. The pro- oncogenic roles of PRMTs such as PRMT1 (upregulated in acute myeloid leukemia, acute megakaryoblastic leukemia) and PRMT5 (upregulated in lymphoma, leukemia, and solid tumors) are well established. Combination therapies of PRMT inhibitors with DNA-damaging agents can promote tumor cell death. Inhibitors for PRMT1 and PRMT5 are presently entering Phase I clinical trials, breaking new grounds for the treatment of solid and hematological malignancies. From a therapeutic perspective, the relevance of each PRMT- regulated process in different cancers need to be elucidated further.
The specificity of these PRMT antibodies, as demonstrated through the data given here, provides researchers with new capabilities to study the epigenetic regulation of histones with increased confidence in their results. To search for PRMT antibodies, please visit thermofisher.com/antibodies.
For research use only. Not for use in diagnostic procedures.
Learn more about ChIP Validation
Additional Antibody Blogs
Let’s get ‘specific’ about the TNFR pathway!
DIY Neurons for antibody validation
Translate to Invitrogen antibodies for your ribosomal protein research!
Drivers of the Chromosomal Passenger Complex
References:
1. Jarrold, James, and Clare C. Davies. “PRMTs and Arginine Methylation: Cancer’s Best-Kept Secret?.” Trends in molecular medicine (2019).
2. Blanc, Roméo S., and Stéphane Richard. “Arginine methylation: the coming of age.” Molecular cell 65.1 (2017): 8-24.
3. Bedford, Mark T., and Steven G. Clarke. “Protein arginine methylation in mammals: who, what, and why.” Molecular cell 33.1 (2009): 1-13.
4. Guccione, Ernesto, and Stéphane Richard. “The regulation, functions and clinical relevance of arginine methylation.” Nat. rev. mol. cell biol 20 (2019): 642-657.
Leave a Reply