Search Thermo Fisher Scientific
Infectious Disease and Microbial Research

Comprehensive Microbial Applications for NGS
From microbial whole genome sequencing using 16S metagenomics to microbiome profiling for human health research to infectious disease research and pathogen identification using targeted sequencing
The need for faster results and better outcomes in disease research has resulted in a migration from traditional molecular biology techniques such as Sanger sequencing, PCR, and culture assays to targeted commensal bacterial species profiling and pathogen identification using next-generation sequencing (NGS).
The evolution of Ion Torrent NGS has enabled researchers to take advantage of increased throughput, higher accuracy, and longer reads to produce rapid and accurate sequencing of microbes with streamlined sample preparation and a simple and optimized data analysis workflow. Ion Torrent sequencing has been critical in facilitating rapid research results from archived samples for disease surveillance and disease etiology determination. Downstream of whole genome microbial and targeted microbiome sequencing, data analysis methods include de novo and reference-guided assembly, antimicrobial resistance detection, and typing of microbial strains.
For targeted sequencing of a specific set of genes, such as 16S rRNA, Ion AmpliSeq technology delivers simple and fast library construction. Based on ultrahigh-multiplex PCR, Ion AmpliSeq technology requires as little as 1 ng of input DNA. Through the use of targeted sequencing, researchers can efficiently identify the microbes within a mixed population without the need to culture samples, perform research on retrospective samples, and discover mutations that may be associated with antibiotic resistance.
Learn about our latest releases:
Ion AmpliSeq SARS-CoV-2 Research Panel
Ion AmpliSeq Microbiome Health Research Kit
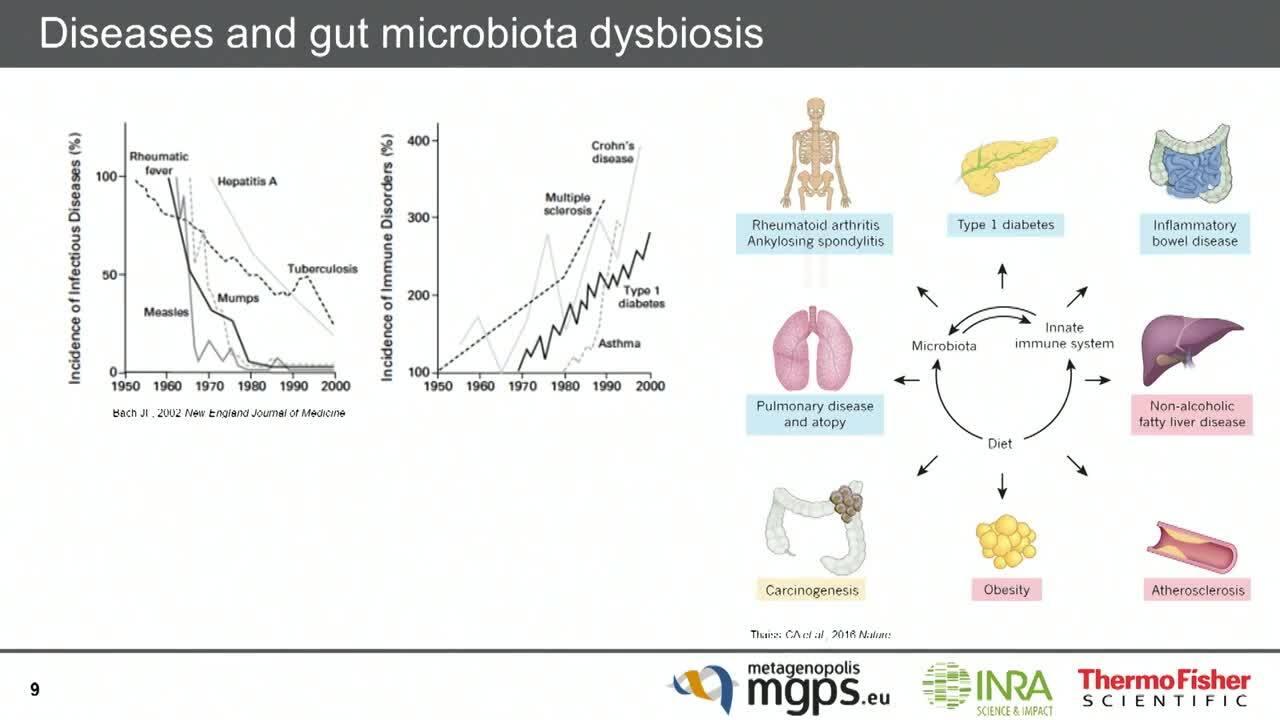
Characterize the gut microbiome for health research

The gut microbiome has emerged as an important biomarker in research into the potential efficacy of immune-modulatory drugs. Using an Ion AmpliSeq highly multiplexed PCR approach enables an efficient and affordable means for conducting extensive analyses of the human microbiome and has applications in the study of phenotypic variability as it relates to disease, emergent resistance mechanisms, and potential future therapies.
Infectious disease research
Bacterial and viral typing
Rapid and affordable bacterial and viral typing research during disease surveillance, using archived samples from investigations and disease etiology studies, such as studies of SARS-CoV-2, MERS, H1N1, and Ebola.
Whole-genome sequencing
16S metagenomics
Deep-coverage whole-genome sequencing of microbial organisms is the ultimate tool for discovering the full range of genetic variations, including SNPs, insertions, deletions, inversions and complex rearrangements. It is used to characterize and discover new organisms or to type specific bacterial and viral organisms.
Targeted sequencing
AmpliSeq panels (microbiome health)
With Ion AmpliSeq technology, you can now perform targeted sequencing of specific genes to identify the microbes within a mixed population, study potential virulence factors and transmission patterns, and discover mutations that may be associated with antibiotic resistance.
Our newest panel, the Ion AmpliSeq Microbiome Health Research Panel, enables robust charactization of bacterial microbes relevant to human diseases at a species level with 100% specificity and sensitivity.
Featured solutions

Infectious disease research
- Ion AmpliSeq SARS-CoV-2 Research Panel
- Ion AmpliSeq Ebola Research Panel
- Ion AmpliSeq Pan-Bacterial Community Panel
- Ion AmpliSeq Antimicrobial Resistance Community Panel
Whole genome sequencing
Targeted sequencing
Order ready-made gene panels or custom design your own at ampliseq.com.
Featured solutions

Infectious disease research
- Ion AmpliSeq SARS-CoV-2 Research Panel
- Ion AmpliSeq Ebola Research Panel
- Ion AmpliSeq Pan-Bacterial Community Panel
- Ion AmpliSeq Antimicrobial Resistance Community Panel
Whole genome sequencing
Targeted sequencing
Order ready-made gene panels or custom design your own at ampliseq.com.
See product selection guide for more details:
The latest infectious disease and microbial research solutions
Ion AmpliSeq SARS-CoV-2 Research Panel
The Ion AmpliSeq SARS-CoV-2 Research Panel is designed to cover more than 99% of the SARS-CoV-2 genome for complete genome sequencing and variant detection for epidemiological studies. Deliver highly accurate coronavirus typing in under a day.
Ion AmpliSeq Microbiome Health Research Kit
The Ion AmpliSeq Microbiome Health Research Kit is a targeted NGS assay for characterizing the human gut microbiome, which has been implicated in research of human disease such as cancer, immunological, and gastrointestinal (GI)
disorder The panels provides the highest species level resolution for the comprehensive profiling of microbial diversity in the human gut microbiome.
Customer videos
What's in our gut effects our health
Hugo Roume of the French National Institute for Agricultural Research discusses the role of the microbiome in human health.
Using Ion 16S sequencing for clinical and environmental research applications
After decades of analyses and hundreds of studies, we’ve learned the diversity of microbes is even greater than imagined. And the diversity of these species can tell us about the well-being of an ecosystem or how their interactions may be linked to diseases such as Inflammatory bowel disease, obesity, and chronic wounds. Hear how Dr. George Watts and Dr. Charles Li are using Ion 16S sequencing in their clinical and environmental research applications.
Microbial sequencing literature & publications
Literature
NEW Application Note: Rapid, automated NGS solution to survey the complete SARS-CoV-2 genome for epidemiological investigation
NEW White Paper: Ion GeneStudio S5 system for multiple SARS-CoV-2 research applications including viral research, host immune response and vaccine research
NEW Ion AmpliSeq Microbiome Health Research Kit
Ion S5 infectious disease application note
Ion AmpliSeq Infectious Disease Research Panels flyer
New 16S flyer
16S algorithm application note
Bacterial typing application note
Poster: Targeted sequencing of 16s RNA Gene to understand diversity and composition of the gut microbiome
Poster: Strain-Typing and Antibiotic Resistance Profiling From Research Samples Using Highly Multiplexed Targeted Library Construction with High Throughput Semi-Conductor Based Sequencing
Poster: Custom next-generation sequencing primer designs for targeted sequencing of multi strain viral targets
Publications
Ion Torrent sequencing is cited in more than 900 peer-reviewed publications on 16S metagenomics, commensal and dysbiosis microbiome for human health research, and microbial genome sequencing.
Bacterial whole genome sequencing and typing
Soni I, et al. (2015). Draft Genome Sequenci of Methicillin-Sensitive Staphylococcus aureus ATCC 29213, ATCC 29213 Genome Announc 3(5): e01095-15. DOI: 10.1128/genomeA.01095-15
Mattos-Guaraldi AL, et al. (2015). Draft Genome Sequence of Corynebacterium striatum1961 BR-RJ/09, a Multidrug-Susceptible Strain Isolated from the Urine of a Hospitalized 37-Year-Old Female Patient
Targeted sequencing
Viral sequencing
16S and metagenome sequencing