Search Thermo Fisher Scientific
Sequence-Specific RNA/DNA Purification
How to capture specific RNA and DNA sequences with streptavidin-coupled Dynabeads
Invitrogen streptavidin-coupled Dynabeads are a robust and versatile tool that can be used to capture specific RNA or DNA sequences and then pull them directly out of solution. These monosized superparamagnetic Dynabeads provide an efficient and solid-phase alternative to nitrocellulose and provide you with an unmatched level of product quality and data consistency. Excellent near-liquid phase reaction kinetics allow for extremely fast protocols. The inherent ease of magnetic handling means that downstream manipulations and buffer changes are as simple as concentrating the bead-bound target at the tube-wall with a magnet and then discarding the supernatant. These beads are compatible with an extremely broad range of sample types including most bodily fluids, crude lysates of plant, animal and microbial origin, as well as purified total RNA or DNA. Since these Dynabeads will only interact with specifically targeted RNA or DNA molecules, upstream purification of total RNA or DNA is almost always an unnecessary step. Learn more about why streptavidin-coupled Dynabeads have been citied in over 30,000 research publications.
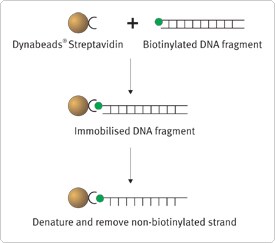
The direct capture procedure involves the immobilization of double-stranded PCR products onto the beads. These are easily converted to single-stranded bead-bound templates which are then used to capture specific RNA or DNA molecules directly from solution.
An alternative indirect capture approach will offer faster reaction kinetics in some cases. This indirect capture procedure allows the target sequence to be captured prior to being immobilized onto the magnetic beads. First, a biotinylated capture-sequence (single-stranded DNA) is incubated with the sample and allowed to hybridize to the targeted RNA or DNA molecules in solution. Streptavidin-coated Dynabeads are then added to the mixture and the hybridized sequences are immobilized onto the Dynabeads via the streptavidin-biotin bond.
The 1 µm Dynabeads MyOne Streptavidin C1 present a very high surface area per mg of beads, enabling high enrichment of low-abundance RNA or DNA. When the goal is to capture nucleic acid from more viscous samples such as cerebrospinal fluid, the larger 2.8 µm sized Dynabeads M-270 Streptavidin are recommended. These Dynabeads (MyOne Streptavidin C1 and M-270 Streptavidin) are optimally designed to have slightly negatively charged surfaces which ensure negligible non-specific binding of non-target nucleic acid sequences.
Examples of applications include: isolation of RNA/DNA infectious agent (1,2,3,4), subtractive hybridization (5,6,7), cDNA selection and enrichment, detection and isolation of mutated sequences (8,9,10), isolation of cell specific transcripts, and mRNA differential display.
Learn more about Nucleic Acid Capture Assays
Selected references
- Meng Q. et al. (2001) Automated multiplex assay system for simultaneous detection of hepatitis B virus DNA, hepatitis C virus RNA and human immunodeficiency virus type 1 RNA. J.Clin.Microbiol. 39(8):2937-2945.
- Stevens SJC. et al. (1999) Monitoring of Epstein-Barr virus DNA load in peripheral blood by quantitative competitive PCR. J.Clin. Microbiol. 37:2852-2857.
- Mangiapan G. et al. (1996) Sequence capture-PCR improves detection of mycobacterial DNA in clinical specimens. J. Clin. Microbiol. 34(5):1209-1215.
- Shuber AP. et.al. (2002) Accurate, noninvasive detection of Helicobacter pylori DNA from stool samples: potential usefulness for monitoring treatment. J. Clin. Microbiol. 40(1):262-264.
- Hansen-Hagge TE. et.al. (2001) Identification of sample-specific sequences in mammalian cDNA and genomic DNA by the novel ligation-mediated subtraction (LIMES). Nucl. Acids Res. 29(4):e20.
- Pradel N. et.al. (2002) Genomic subtraction to identify and characterize sequences of Shiga toxin-producing Escherichia coli O91:H21. Appl. Env. Microbiol. 68(5):2316-2325.
- Laveder P. et.al. (2002) A two-step strategy for constructing specifically self-subtracted cDNA libraries. Nucleic Acids Res. 30(9):e38.
- Lindblad-Toh K.et.al. (2000) Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nature Genetics. 24:381-386.
- Miyashiro I. et.al. (2001) Molecular strategy for detecting metastatic cancers with use of multiple tumor-specific MAGE-A genes. Clin.Chem. 47(3):505-512.
- Dong SM. et.al. (2001) Detection of colorectal cancer in stool with the use of multiple genetic targets. J Natl. Cancer Inst. 93(11):858-865.