Search Thermo Fisher Scientific
Five Steps to Fast RT-PCR
In reverse transcription polymerase chain reaction (RT-PCR), an RNA population is converted to cDNA by reverse transcription (RT), and then the cDNA is amplified by the polymerase chain reaction (PCR). Common applications of RT-PCR include detection of expressed genes, examination of transcript variants, and generation of cDNA templates for cloning and sequencing. Since reverse transcription provides cDNA templates for PCR amplification and downstream experiments, it is one of the most critical steps for experimental success. Explore the following five steps to fast RT-PCR and gain a better understanding of the RT-PCR procedure.
Video: One-step vs two-step RT-PCR—which one is right for your experiment?
Video: How to get high specificity in one-step RT-PCR results
Determine which RT-PCR procedure to use
In performing RT-PCR, one-step and two-step methods are the two common approaches, each with its own advantages and disadvantages. One-step RT-PCR combines first-strand cDNA synthesis (RT) and subsequent PCR in a single reaction tube. However, two-step RT-PCR entails two separate reactions, beginning with first-strand cDNA synthesis, followed by amplification of a portion of the resulting cDNA by PCR in a separate tube.
Figure 1. One-step and two-step RT-PCR.
One-step RT-PCR | Two-step RT-PCR | |
|---|---|---|
| Setup | Combine reaction under conditions that support both reverse transcription and PCR | Separate optimized reaction for reverse transcription and PCR |
| Primers | Gene-specific | Oligo (dT), random hexamers or gene-specific primers |
| Ideal use | Analysis of one or two genes, high-throughput platforms | Analysis of multiple genes |
| Advantage |
|
|
For a faster and simpler workflow, a one-step approach is often preferred over the separate two steps.
Learn more: One-step vs two-step RT-PCR
Prepare sample for RT-PCR
Total RNA is routinely used in one-step RT-PCR. Maintaining RNA integrity is critical and requires special precautions during extraction, processing, storage, and experimental use. The main goals of isolation workflows are to stabilize RNA molecules, inhibiting RNases, and maximizing yield with proper storage and extraction methods. Optimal purification methods remove endogenous compounds, like complex polysaccharides and humic acid from plant tissues that interfere with enzyme activity, and common inhibitors of reverse transcriptases, such as salts, metal ions, ethanol, and phenol. Once purified, RNA should be stored at –80°C with minimal freeze-thaw cycles.
Explore: RNA isolation
Inhibitors in reverse transcription reactions
Engineered reverse transcriptases are able to withstand common inhibitors of reverse transcriptase and PCR, such as co-purified compounds from biological samples or reagents used for RNA purification. This exceptional robustness makes reverse transcription less dependent on RNA sample purity to achieve reliable results.

Figure 2. Resistance to inhibitors. Detection of a 1 kb RNA target from 100 ng UHRR using the SuperScript IV UniPrime One-Step RT-PCR System or other one-step RT-PCR kits in reaction mixtures containing: (1) no inhibitor, (2) Xylan (2.5 μg/μL), (3) Humic Acid (15 ng/μL), (4) SDS (0.013%), (5) Trizol (1.2%), or (6) LiCl (2.5 μg/μL).
Explore: Reverse transcription products
Design primers for one-step PCR
In one-step RT-PCR it is recommended to use gene-specific primers (GSPs). Oligo (dT) or random primers are not recommended, because non-specific products can be generated, thereby reducing the amount of target RT-PCR products. Designing good GSPs is the first step toward success of a one-step RT-PCR experiment.
Primer design tips
- Design primers that anneal to the mRNA sequence in exons on both sides of an intron or exon/exon boundary, to allow differentiation between the amplified cDNA and potential contaminating genomic DNA
- If this approach is not feasible, consider using the next step—genomic DNA removal with ezDNase Enzyme
- Ensure primers are not self- complementary or are not complementary to each other at the 3’ end
- A final concentration of 0.5 uM for each primer is recommended, but further optimization may be necessary
- To calculate primer Tm and estimate appropriate annealing temperatures for PCR, use a Tm calculator
The Invitrogen SuperScript IV UniPrime One-Step RT-PCR System has a universal primer annealing temperature of 60ºC, which eliminates the need to calculate the melting temperature (Tm) of each primer set. Primer annealing is a critical step in PCR, and using a universal annealing temperature simplifies the process of PCR optimization, which can be tedious and time-consuming.
Remove genomic DNA from the RNA sample
Since small amounts of genomic DNA can occasionally contaminate purified RNA, there are three common approaches to help minimize the impact of contaminating genomic DNA in a reaction.
- Design primers across exon-exon junctions to prevent genomic DNA amplification
- Use control reactions without reverse transcriptase to monitor genomic DNA contamination
- Remove genomic DNA by DNase treatment
Genomic DNA removal with conventional DNases may cause different levels of RNA loss or damage, leading to inconsistent results. A newer generation DNase—Invitrogen ezDNase Enzyme significantly reduces the possibility of cDNA synthesis being compromised due to DNase I treatment. ezDNase Enzyme is a recombinant double-strand-specific DNase for the fast removal of contaminating genomic DNA from RNA preparations with features include:
- Efficient and fast genomic DNA removal
- Highly specific—no impact on RNA, cDNA, or primers in RT reactions
ezDNase Enzyme's high specificity for double-stranded DNA enables efficient and fast genomic DNA removal without reduction in the quality or quantity of RNA or single-stranded DNA present in the reaction such as cDNA and primers. ezDNase Enzyme is heat-labile and so can be easily deactivated by heat treatment at moderate temperature (55°C). These features make ezDNase an exceptional choice for genomic DNA removal prior to reverse transcription reactions.
Figure 3. gDNA removal procedures: DNase I vs. Invitrogen ezDNase Enzyme. Compared to DNase I, ezDNase Enzyme offers a shorter workflow, simpler procedure, and less RNA damage. Inactivation of ezDNase Enzyme prior to reverse transcription is optional since the enzyme does not cleave primers, ssRNA, or cDNA:RNA complexes.
Explore: Invitrogen ezDNase Enzyme
Perform one-step RT-PCR
Once steps one through four are achieved, it is time to perform one-step RT-PCR. RT-PCR involves two main reactions: first-strand cDNA synthesis and amplification of cDNA. In one-step RT-PCR, synthesis of cDNA and the PCR reaction are combined under conditions that support both reverse transcription and PCR. This is performed in a single reaction tube in a common reaction buffer as flows:
- Thaw reagents
- Prepare RT-PCR reaction mix
- Program thermocycler
- Run thermocycler
- Analyze with gel electrophoresis
For a quick and convenient one-step RT-PCR process consider the Invitrogen SuperScript IV UniPrime One-Step RT-PCR System. This system offers exceptional performance and ease of use by combining the highly processive SuperScript IV Reverse Transcriptase with a novel UniPrime RT-PCR Master Mix.
Explore: One-step RT-PCR systems
Exceptional performance with analytical sensitivity down to 0.01 pg of RNA

Figure 4. Target detection from low amounts of input RNA. Amplification of 0.43 kb fragment from serial dilution from 1 μg to 0.01 pg of UHRR with SuperScript IV UniPrime One-Step RT-PCR System. The molecular weight marker is Thermo Scientific GeneRuler 100 bp Plus DNA Ladder, ready-to-use.
Target length up to 13.8 kb
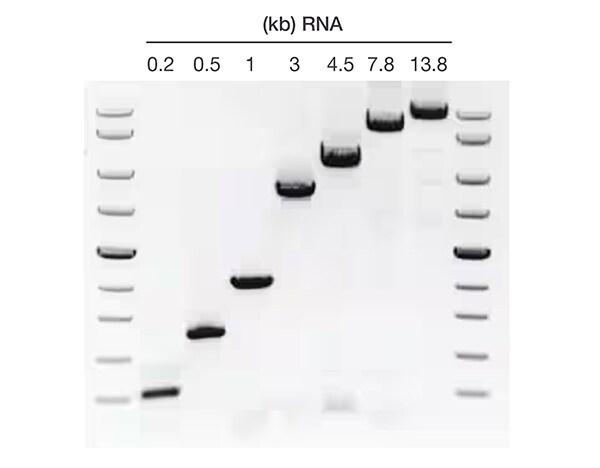
Figure 5. Versatility across a broad range of target lengths. Detection of human RNA fragments ranging from 0.2 to 13.8 kb with the SuperScript IV One-Step RT-PCR System.
High analytical specificity with yield with the shortest protocol

Figure 6. Amplification of long targets with high specificity in significantly shorter times. Detection of 7.8 kb target from total HeLa RNA using SuperScript IV One-Step RT-PCR System, SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity, and supplier QI, N, QU, T, R, and BL one-step RT-PCR products. Reactions were performed according to suppliers’ recommendations. Total reaction times for one-step RT-PCR are indicated in hours:minutes. The RNA target failed to amplify with products from suppliers QI and BL.
Explore: One-step RT-PCR systems
Learn more: Reverse transcription troubleshooting guide
Explore: Five step workflows
Determine which RT-PCR procedure to use
In performing RT-PCR, one-step and two-step methods are the two common approaches, each with its own advantages and disadvantages. One-step RT-PCR combines first-strand cDNA synthesis (RT) and subsequent PCR in a single reaction tube. However, two-step RT-PCR entails two separate reactions, beginning with first-strand cDNA synthesis, followed by amplification of a portion of the resulting cDNA by PCR in a separate tube.
Figure 1. One-step and two-step RT-PCR.
One-step RT-PCR | Two-step RT-PCR | |
|---|---|---|
| Setup | Combine reaction under conditions that support both reverse transcription and PCR | Separate optimized reaction for reverse transcription and PCR |
| Primers | Gene-specific | Oligo (dT), random hexamers or gene-specific primers |
| Ideal use | Analysis of one or two genes, high-throughput platforms | Analysis of multiple genes |
| Advantage |
|
|
For a faster and simpler workflow, a one-step approach is often preferred over the separate two steps.
Learn more: One-step vs two-step RT-PCR
Prepare sample for RT-PCR
Total RNA is routinely used in one-step RT-PCR. Maintaining RNA integrity is critical and requires special precautions during extraction, processing, storage, and experimental use. The main goals of isolation workflows are to stabilize RNA molecules, inhibiting RNases, and maximizing yield with proper storage and extraction methods. Optimal purification methods remove endogenous compounds, like complex polysaccharides and humic acid from plant tissues that interfere with enzyme activity, and common inhibitors of reverse transcriptases, such as salts, metal ions, ethanol, and phenol. Once purified, RNA should be stored at –80°C with minimal freeze-thaw cycles.
Explore: RNA isolation
Inhibitors in reverse transcription reactions
Engineered reverse transcriptases are able to withstand common inhibitors of reverse transcriptase and PCR, such as co-purified compounds from biological samples or reagents used for RNA purification. This exceptional robustness makes reverse transcription less dependent on RNA sample purity to achieve reliable results.

Figure 2. Resistance to inhibitors. Detection of a 1 kb RNA target from 100 ng UHRR using the SuperScript IV UniPrime One-Step RT-PCR System or other one-step RT-PCR kits in reaction mixtures containing: (1) no inhibitor, (2) Xylan (2.5 μg/μL), (3) Humic Acid (15 ng/μL), (4) SDS (0.013%), (5) Trizol (1.2%), or (6) LiCl (2.5 μg/μL).
Explore: Reverse transcription products
Design primers for one-step PCR
In one-step RT-PCR it is recommended to use gene-specific primers (GSPs). Oligo (dT) or random primers are not recommended, because non-specific products can be generated, thereby reducing the amount of target RT-PCR products. Designing good GSPs is the first step toward success of a one-step RT-PCR experiment.
Primer design tips
- Design primers that anneal to the mRNA sequence in exons on both sides of an intron or exon/exon boundary, to allow differentiation between the amplified cDNA and potential contaminating genomic DNA
- If this approach is not feasible, consider using the next step—genomic DNA removal with ezDNase Enzyme
- Ensure primers are not self- complementary or are not complementary to each other at the 3’ end
- A final concentration of 0.5 uM for each primer is recommended, but further optimization may be necessary
- To calculate primer Tm and estimate appropriate annealing temperatures for PCR, use a Tm calculator
The Invitrogen SuperScript IV UniPrime One-Step RT-PCR System has a universal primer annealing temperature of 60ºC, which eliminates the need to calculate the melting temperature (Tm) of each primer set. Primer annealing is a critical step in PCR, and using a universal annealing temperature simplifies the process of PCR optimization, which can be tedious and time-consuming.
Remove genomic DNA from the RNA sample
Since small amounts of genomic DNA can occasionally contaminate purified RNA, there are three common approaches to help minimize the impact of contaminating genomic DNA in a reaction.
- Design primers across exon-exon junctions to prevent genomic DNA amplification
- Use control reactions without reverse transcriptase to monitor genomic DNA contamination
- Remove genomic DNA by DNase treatment
Genomic DNA removal with conventional DNases may cause different levels of RNA loss or damage, leading to inconsistent results. A newer generation DNase—Invitrogen ezDNase Enzyme significantly reduces the possibility of cDNA synthesis being compromised due to DNase I treatment. ezDNase Enzyme is a recombinant double-strand-specific DNase for the fast removal of contaminating genomic DNA from RNA preparations with features include:
- Efficient and fast genomic DNA removal
- Highly specific—no impact on RNA, cDNA, or primers in RT reactions
ezDNase Enzyme's high specificity for double-stranded DNA enables efficient and fast genomic DNA removal without reduction in the quality or quantity of RNA or single-stranded DNA present in the reaction such as cDNA and primers. ezDNase Enzyme is heat-labile and so can be easily deactivated by heat treatment at moderate temperature (55°C). These features make ezDNase an exceptional choice for genomic DNA removal prior to reverse transcription reactions.
Figure 3. gDNA removal procedures: DNase I vs. Invitrogen ezDNase Enzyme. Compared to DNase I, ezDNase Enzyme offers a shorter workflow, simpler procedure, and less RNA damage. Inactivation of ezDNase Enzyme prior to reverse transcription is optional since the enzyme does not cleave primers, ssRNA, or cDNA:RNA complexes.
Explore: Invitrogen ezDNase Enzyme
Perform one-step RT-PCR
Once steps one through four are achieved, it is time to perform one-step RT-PCR. RT-PCR involves two main reactions: first-strand cDNA synthesis and amplification of cDNA. In one-step RT-PCR, synthesis of cDNA and the PCR reaction are combined under conditions that support both reverse transcription and PCR. This is performed in a single reaction tube in a common reaction buffer as flows:
- Thaw reagents
- Prepare RT-PCR reaction mix
- Program thermocycler
- Run thermocycler
- Analyze with gel electrophoresis
For a quick and convenient one-step RT-PCR process consider the Invitrogen SuperScript IV UniPrime One-Step RT-PCR System. This system offers exceptional performance and ease of use by combining the highly processive SuperScript IV Reverse Transcriptase with a novel UniPrime RT-PCR Master Mix.
Explore: One-step RT-PCR systems
Exceptional performance with analytical sensitivity down to 0.01 pg of RNA

Figure 4. Target detection from low amounts of input RNA. Amplification of 0.43 kb fragment from serial dilution from 1 μg to 0.01 pg of UHRR with SuperScript IV UniPrime One-Step RT-PCR System. The molecular weight marker is Thermo Scientific GeneRuler 100 bp Plus DNA Ladder, ready-to-use.
Target length up to 13.8 kb

Figure 5. Versatility across a broad range of target lengths. Detection of human RNA fragments ranging from 0.2 to 13.8 kb with the SuperScript IV One-Step RT-PCR System.
High analytical specificity with yield with the shortest protocol

Figure 6. Amplification of long targets with high specificity in significantly shorter times. Detection of 7.8 kb target from total HeLa RNA using SuperScript IV One-Step RT-PCR System, SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity, and supplier QI, N, QU, T, R, and BL one-step RT-PCR products. Reactions were performed according to suppliers’ recommendations. Total reaction times for one-step RT-PCR are indicated in hours:minutes. The RNA target failed to amplify with products from suppliers QI and BL.
Explore: One-step RT-PCR systems
Learn more: Reverse transcription troubleshooting guide
Explore: Five step workflows
For Research Use Only. Not for use in diagnostic procedures.

