Search Thermo Fisher Scientific
Molecular Insights on Microbial Challenges

Microbial identification, detection, and role in disease
Genetic analysis technologies have revolutionized microbiology and infectious disease research, expanding our knowledge of microbial diversity and its implication in mechanisms of pathology. Thermo Fisher Scientific offers a broad range of genetic analysis technologies to identify, detect, and characterize microbes faster than traditional microbiological culture techniques.
Our optimized platforms for next-generation sequencing (NGS), Sanger sequencing using capillary electrophoresis, real-time PCR, and microarrays offer highly effective approaches to analyzing viruses, bacteria, and fungi. These technologies are also leveraged to study human response to microorganisms. Ultimately, research leading to better understanding of host–pathogen interactions can translate into better prevention and treatment of infectious diseases.
The complete solution for reliable identification of genotypic microbes
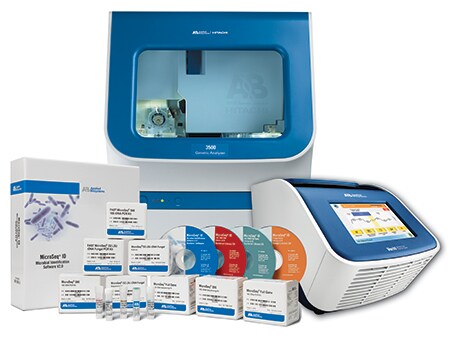
Enable unparalleled accuracy and time-to-results for bacterial and fungal identifications with Applied Biosystems 3500 Series genetic analyzers. Built on years of molecular expertise, this easy-to-use platform enables accurate genotypic bacterial identification based on the 16S rRNA gene, and accurate genotypic fungal identification based on the D2 region of the 26S rRNA gene. With an easy workflow and high-throughput capabilities, you can expect accurate results in less than five hours.
Learn more about Applied Biosystems MicroSEQ Rapid Microbial Identification System
Increased speed and accuracy in urinary tract pathogen detection

Our newly released real-time PCR solution for urinary tract microbiota (UTM) detection offers a sample-to- answer workflow in just five hours. Additionally, it increases the accuracy in detecting urinary tract pathogens by 25% when compared to culture-based methods. Clinical verification data demonstrates that when compared with urine specimen culture, real-time PCR detected 97% of samples where urinary tract pathogens were present, whereas culture detected only 71% of cases where pathogens were present.
Learn more about our real-time PCR solution for detecting urinary tract microbiota (UTM)
Free on-demand webinars
Impact of malaria on biological aging
In the webinar “Infectious diseases accelerate allostatic load and biological aging,” Dr. Muhammad Asghar of the Karolinska Institute Stockholm presents the cellular mechanisms associated with aging, and how real-time PCR was used to study the impact of chronic asymptomatic malarial infections on telomere degradation.

Human transcriptome profiling from fecal samples
Transcriptome profiling from clinical research samples is commonly used for disease biomarker discovery. However, blood, FFPE, and feces are challenging samples to work with due to limited extractable RNA. In this webinar, Dr. Mark Manary and Dr. Sean Diehl describe how to measure the human transcriptome from feces (>95% bacterial RNA) with high-performance array technology. They identify biomarkers for environmental enteric dysfunction, a major cause of stunting in the developing world. Overall, this methodology can benefit researchers investigating the effects of the microbiome or microbial infections on gut disease.

Analytical validation consulting service aids launch of new laboratory-developed test for women’s health.
For Research Use Only. Not for use in diagnostic procedures.