Search Thermo Fisher Scientific
- Order Status
- Quick Order
-
Don't have an account ? Create Account
Search Thermo Fisher Scientific
Gene fusions and their importance in cancer research
Chromosome aberrations, in particular translocations and their corresponding gene fusions, have been shown to play an important role in the initial steps of tumorigenesis. The identification of gene fusions shows promise for playing an important role in personalized cancer treatment decisions in the future. Many rare gene fusion events have been identified in fresh-frozen solid tumors from common cancers, by next-generation sequencing technology. However, the ability to detect transcripts from gene fusions in RNA isolated from formalin-fixed, paraffin-embedded (FFPE) tumor tissues has been limited, due to the low complexity of FFPE libraries and the associated bioinformatics challenges.
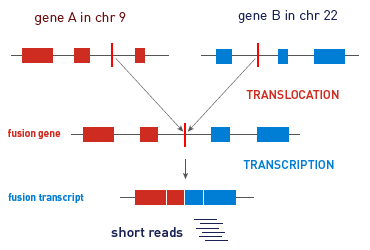
In collaboration with the OncoNetwork Consortium, we have developed a research method that simplifies the detection of chromosomal translocations. Designed for simplicity and efficiency and to work with limited sample input, this method delivers sensitive, reliable results with ultralow input of FFPE RNA, enabling you to detect fusion transcripts in less than 1% tumor RNA in the presence of 99% normal RNA.
Based on Ion AmpliSeq™ technology, our fusion detection workflow is simple, with reduced cost and complexity compared with alternative fusion detection methods such as whole-transcriptome sequencing and FISH. Based on simple PCR, Ion AmpliSeq™ technology uses high-multiplexing capabilities to enable you to find and view many fusions in a single run. This targeted sequencing approach focuses the sequencing on fusion junctions, increasing the depth of sequencing on informative regions of fusion transcripts. This enables higher sensitivity of detection and greater sequencing efficiency to save both time and money.
"With these two panels, we finally have a tool to determine the main mutations involved in lung cancer research starting from a single archived tumor sample with limited amount of material."
Nicola Normanno, MD, PhD
Director, Research Department Centro di Ricerche Oncologiche di Mercogliano
Mercogliano (AV) and INT-Fondazione Pascale, Naples, Italy
OncoNetwork Consortia Member
Our fusion detection solution starts with only 10 ng of FFPE RNA, resulting in sensitive fusion detection with the Ion PGM™ System. This system is integrated with Ion Reporter™ Software, which includes easy-to-use fusion visualization tools, fusion classification, and gene expression details.
Built to support RNA fusion data analysis, Ion Reporter Software v4.2 enables push-button fusion data analysis that delivers multi-sample visualization and read-level visualization of gene fusions.
This software also enables the creation of heat maps to show exon–exon boundaries between gene partners, to show chimeric RNA covering these exons.
For Research Use Only. Not for use in diagnostic procedures.



