Search Thermo Fisher Scientific
GeneOptimizer Process for Successful Gene Optimization

The power of custom gene synthesis is in the ability to design your DNA according to your needs and without the constraints of traditional cloning. Equally important to most researchers, however, is obtaining high yields of mRNA and ultimately protein from synthetic genes. We developed GeneArt GeneOptimizer software to maximize expression of synthetic genes in all available expression systems. Sequence optimization is optional with every GeneArt project.
We do more than codon optimization
What is gene optimization?
Gene optimization takes advantage of the degeneracy of the genetic code. Because of degeneracy, one protein can be encoded by many alternative nucleic acid sequences. Codon preference (codon usage bias) differs in each organism, and it can create challenges for expressing recombinant proteins in heterologous expression systems, resulting in low and unreliable expression. This may also be true for autologous expression, since wild type sequences are not necessarily optimized for expression yield but also for degradation, regulation, and other properties.
However, codon optimization is not the only relevant factor for efficient protein expression.
Our GeneOptimizer algorithm enables true multiparametric optimization, dealing with a large number of sequence-related parameters involved in different aspects of gene expression, such as transcription, splicing, translation, and mRNA degradation. It considers all relevant optimization parameters in a single operation and delivers a DNA sequence configured with your specifications, optimized for maximum performance in your system (Table 1).
Rationally weighing the combination of the optimization parameters (e.g., codon adaptation, mRNA de novo synthesis and stability, transcription and translation efficiency) is important in order to achieve the most efficient expression of a given protein.

Figure 1. Schematic representation of protein expression.
Table 1. Parameters that influence protein expression.
| Transcriptional level | mRNA level | Translational level |
|---|---|---|
|
|
|
One further advantage of synthetic sequences is that you won’t be dependent on available DNA templates, and you can design your sequence exactly according to your requirements. Add or remove restriction sites, start/stop codons, tags, and further motifs as needed.
Sequence optimization using the GeneOptimizer software is included as an optional step with all GeneArt Gene Synthesis and DNA fragments services. To take advantage of this service, select your expression host when setting up a request using our online customer portal. The online software will then guide you through the project setup process, including sequence optimization. The resulting optimized performance, described below, is just one additional added value of GeneArt DNA.
How does it work?
Gene optimization with GeneOptimizer technology is easily performed within a few minutes using our online customer portal. Design your synthetic gene by uploading your sequence, selecting your expression system, and specifying your cloning vector and your sequence details (including open reading frames, untranslated regions, and cloning sites). Once you submit your request, the GeneOptimizer software generates the DNA sequence that best suits your research requirements, based on consideration of all parameters that are relevant for the given host organism plus your individual sequence requirements.
Proof that it works
We have conducted several internal studies, and many customers have reported independently that GeneArt codon and sequence optimization results in higher protein expression without losing protein function.
In a first-of-its-kind study [1], five important protein classes were selected for optimization: protein kinases, transcription factors, ribosomal proteins, cytokines, and membrane proteins. Then 50 human genes were chosen from the NCBI database to represent the five protein classes. The selected genes were individually optimized using the GeneOptimizer algorithm [2]. For comparison, the corresponding wild type genes were subcloned using native sequences available from the NCBI database. Each gene was then expressed in triplicate in HEK293T cells. Following optimization, the 50 genes all showed reliable expression and 86% exhibited elevated expression (example in Figure 2). Further analysis showed no detrimental effect on protein solubility, and functionality was unaltered, as demonstrated for JNK1, JNK3, and CDC2 (data not shown).
Using the GeneOptimizer algorithm, in this study:
- 86% of optimized genes showed significantly increased protein expression
- Protein yields increased up to 15-fold with optimized genes
- 100% of optimized genes were expressed, versus 88% of wild type genes

Figure 2. Comparative expression analysis of wild type vs. optimized genes representing different protein classes. (A) Cell culture supernatants (for secreted proteins) or cell lysates (all other proteins) were analyzed by western blot using an anti-His antibody. One example of each protein class is shown. A 60 kDa protein used to standardize protein amount is visible, including in the empty vector negative controls. Left of each image: molecular mass values in kDa. Right of each image: identifiers for specific protein bands. (B) Relative expression levels were derived for wild type or optimized constructs (mean of three independent transfections). The fold increase in expression for the optimized construct is indicated for each protein. There was no detectable expression for IL-2 using the wild type construct. (Figure adapted from Fath et al., 2011 [1]).
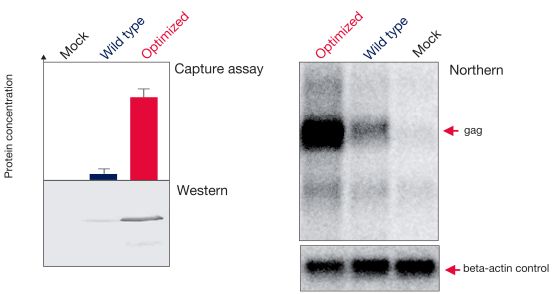
Figure 3 illustrates another example of observed increases in both mRNA and protein yields of the HIV gag protein after sequence optimization using GeneOptimizer software.
To demonstrate the value of the GeneOptimizer software, we compared protein expression of sequences optimized by different vendors. Genes for three different human kinases were optimized and synthesized in-house or similarly optimized and synthesized by five different competitors. Triplicate expression studies in HEK293 cells showed not only that GeneArt optimization raises expression over wild type genes, but also that it performs better than any of the five competitors’ optimization algorithms in every case (Figure 4).

Figure 4. Expression levels from wild type genes and the same genes optimized by GeneArt technology and five competitors. Relative protein expression values are normalized to the respective GeneArt sequence.
References
- Fath et al.: Multiparameter RNA and Codon Optimization
- Raab et al.: The GeneOptimizer algorithm: using a sliding window approach to cope with the vast sequence space in multiparameter DNA sequence optimization
- Graf et al.: Codon-Optimized Genes that Enable Increased Heterologous Expression in Mammalian Cells and Elicit Efficient Immune Responses in Mice after Vaccination of Naked DNA
Request more information about GeneArt optimization and our gene synthesis service online or send us an email to geneartsupport@thermofisher.com
Resources
Literature
References
Videos
Related products and services
For Research Use Only. Not for use in diagnostic procedures.