Search Thermo Fisher Scientific
Educational Resources for TaqMan® MicroRNA Assays

MicroRNA (miRNA) and other small ncRNAs provide a natural gene silencing mechanism. They play a central role in controlling how the genome is regulated, and how traits are passed on or eliminated by environmental and genetic factors. We offer TaqMan® Assays and other tools specifically for miRNA and ncRNA research, from isolation through discovery, profiling, quantitation, validation, and functional analysis. Learn more about TaqMan® MicroRNA Assays and related products below.
Featured TaqMan MicroRNA® Assay resources
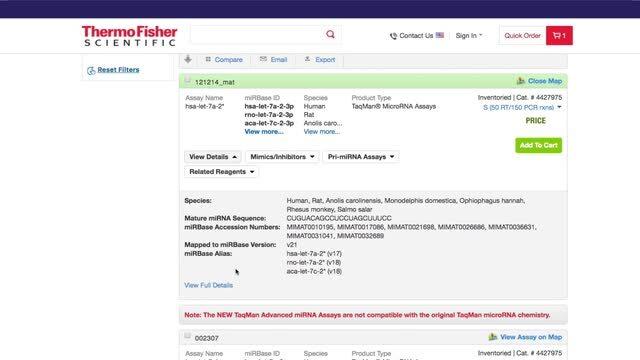
15K+ microRNA assays based on miRBase v21, including associated miRBase ID, accession number and alias.
Understanding miRBase nomenclature, normalizing results, searching and ordering online, and more.
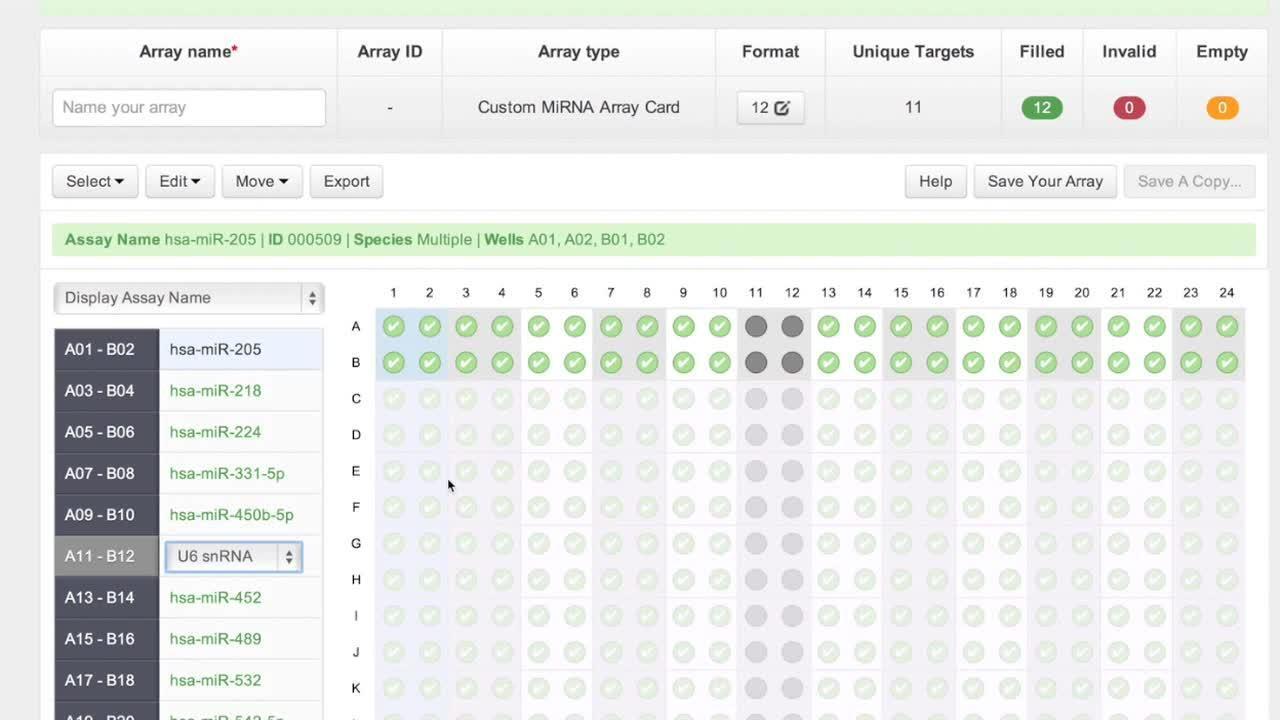
Creating Custom RT and Preamplification Pools
Download the protocol for combining up to 96 microRNA RT primers together for multiplexed reverse transcription.
MicroRNA analysis: From discovery to functional solutions with Life Technologies
Explore our solutions for microRNA analysis, from discovery to functional analysis. Download our microRNA resource guide and learn about innovative tools for microRNA research.
Which TaqMan® microRNA or noncoding RNA assay/array is right for you?
TaqMan® MicroRNA Assays are available in these formats:
- Single tubes are a convenient all-in-one tube format for quantitative microRNA and noncoding RNA (ncRNA) studies
- 384-well microfluidic cards are ideal for profiling studies of miRNA only on our ViiA™ 7 and 7900HT Fast Real-Time PCR Systems
- OpenArray® plates are for use with the OpenArray® Real-Time PCR System to meet high-throughput profiling demands
Choose your TaqMan® Assay/Array based on your research needs:
| What assay format do you want? | What type of research are you doing? | Use this TaqMan® Assay/Array |
|---|---|---|
| Single tube | Target microRNA analysis | |
| Pri-miRNA (human/rodent) analysis | ||
| Long noncoding RNA (human/rodent) analysis | ||
| Any of the above, requiring custom-designed assays | ||
| 384-well microfluidic card | Comprehensive coverage of most relevant microRNA (miRBase v20) using limited samples or material (human/rodent) | TaqMan® Array MicroRNA Cards |
| Profiling selected microRNAs (using predesigned assays) | Custom TaqMan® Array MicroRNA Cards | |
| OpenArray® plate | Large-scale profiling (human/rodent) | TaqMan® OpenArray® MicroRNA Panels |
TaqMan® MicroRNA Assays featured videos
Dr. Leendert H. J. Looijenga of the Erasmus Medical Center in the Netherlands discusses his research focused on DNA, mRNA, miRNA, and protein levels to identify the earliest changes in the development of cancer. Learn how Dr. Looijenga uses Life Technologies™ products to detect target miRNA in body fluids.
Expression analysis of both mRNA and miRNA on the same TaqMan® Array card, Dr. Malte Bucholz
Dr. Malte Buchholz of the Clinic for Gastroenterology at the University of Marburg, Germany, discusses his pancreatic cancer research using the simultaneous measurement and analysis of mRNA and miRNA in the same sample with a single Custom TaqMan® Array card.
TaqMan® MicroRNA Assays key citations
Chang KH et al. (2010) MicroRNA expression profiling to identify and validate reference genes for relative quantification in colorectal cancer.BMC Cancer 10:173; doi: 10.1186/1471-2407-10-17.
Das S et al. (2013) Modulation of neuroblastoma disease pathogenesis by an extensive network of epigenetically regulated microRNAs.Oncogene 32(24):2927-2936; doi: 10.1038/onc.2012.
Di Leva G et al. (2013) Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status.PLoS Genet 9(3); doi: 10.1371/journal.pgen.1003311.
Francia S et al. (2012) Site-specific DICER and DROSHA RNA products control the DNA damage response. Nature 488(7410): 231–235; doi: 10.1038/nature11179.
Havelange V et al. (2011) Functional implications of microRNAs in acute myeloid leukemia by integrating microRNA and mRNA expression profiling. Cancer 117(20), doi: 10.1002/cncr.26096.
Hammell CM, Karp X, Ambros V (2009) A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc Natl Acad Sci U S A 106(44): 18668–18673; doi: 10.1073/pnas.0908131106.
Hennessey PT et al. (2012) Serum microRNA biomarkers for detection of non-small cell lung cancer. PLoS One 7(2); doi: 10.1371/journal.pone.0032307.
Hudson RS et al. (2012) MicroRNA-1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic Acids Res 40(8): 3689–3703; doi: 10.1093/nar/gkr1222.
Karp X et al. (2011) Effect of life history on microRNA expression during C. elegans development. RNA 17(4): 639–651; doi: 10.1261/rna.2310111. 17(4): 639–651. doi: 10.1261/rna.2310111. Epub 2011 Feb 22.
Langevin SM et al. (2011) MicroRNA-137 promoter methylation is associated with poorer overall survival in patients with squamous cell carcinoma of the head and neck. Cancer 117(7): 1454–1462; doi: 10.1002/cncr.25689.
Nazari-Jahantigh M et al. (2012) MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J Clin Invest 122(11): 4190–4202; doi: 10.1172/JCI61716. Epub 2012 Oct 8.
Patterson EE et al. (2011) MicroRNA profiling of adrenocortical tumors reveals miR-483 as a marker of malignancy. Cancer 117(8): 1630–1639; doi: 10.1002/cncr.25724.
Romano G et al. (2012) MiR-494 is regulated by ERK1/2 and modulates TRAIL-induced apoptosis in non–small-cell lung cancer through BIM down-regulation. Proc Natl Acad Sci U S A 109(41): 16570–16575; doi: 10.1073/pnas.1207917109.
Sheinerman KS et al. (2012) Plasma microRNA biomarkers for detection of mild cognitive impairment. Aging 4(9): 590–605.
van Schooneveld E et al. (2012) Expression profiling of cancerous and normal breast tissues identifies microRNAs that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers.Breast Cancer Res. 14(1): R34; doi: 10.1186/bcr3127.
Su X et al. (2010) TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 467(7318): 986–990; doi: 10.1038/nature09459.
Tili E et al. (2011) Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc Natl Acad Sci U S A 108(12): 4908–4913; doi: 10.1073/pnas.
Wang W et al. (2012) Identification of deregulated miRNAs and their targets in hepatitis B virus-associated hepatocellular carcinomaWorld J Gastroenterol 18(38): 5442–5453; doi: 10.3748/wjg.v18.i38.5442.
Yoshizawa S et al. (2012) Downregulated plasma miR-92a levels have clinical impact on multiple myeloma and related disorders. Blood Cancer J 2(1): e53; doi: 10.1038/bcj.2011.51.
Yu J et al. (2012) MicroRNA alterations of pancreatic intraepithelial neoplasms (PanINs). Clin Cancer Res 18(4): 981–992; doi: 10.1158/1078-0432.CCR-11-2347.
Zisoulis DG, Kai ZS, Chang RK, Pasquinelli AE (2012) Auto-regulation of miRNA biogenesis by let-7 and Argonaute. Nature 486(7404): 541–544; doi: 10.1038/nature11134.
TaqMan® MicroRNA Assays documents & resources
Browse the tabs below to find application notes, videos, and protocols for TaqMan® MicroRNA Assays.
Product bulletins
TaqMan® MicroRNA Assays and arrays
TaqMan® Pri-miRNA Assays: detect the origins of microRNA expression
TaqMan® miRNA ABC Purification Kit
Brochures
Additional resources
Complete list of TaqMan® MicroRNA Assays
QRC: TaqMan® Small RNA Assays
Design process for small RNA assays
Customer focus story: TaqMan® MicroRNA Assays help researchers validate miRNA markers for forensic body fluid identification
Application notes
TaqMan® Array Cards for combined mRNA and miRNA analysis
Endogenous controls for real-time quantitation of miRNA using TaqMan® MicroRNA assays
High correlation of miRNA quantitation data from matched FFPE and snap-frozen tissues using TaqMan® MicroRNA Assays
Optimized protocol with low sample input for profiling human microRNA using the OpenArray® platform
Optimized blood plasma protocol for profiling human miRNAs using the OpenArray® Real-Time PCR System
Posters
MicroRNA quantitation by real-time PCR
TaqMan®-based miRNA profiles classify embryonic stem and differentiated cells
Custom RT and PreAmp pools using TaqMan® miRNA Assays
Product bulletins
TaqMan® MicroRNA Assays and arrays
TaqMan® Pri-miRNA Assays: detect the origins of microRNA expression
TaqMan® miRNA ABC Purification Kit
Brochures
Additional resources
Complete list of TaqMan® MicroRNA Assays
QRC: TaqMan® Small RNA Assays
Design process for small RNA assays
Customer focus story: TaqMan® MicroRNA Assays help researchers validate miRNA markers for forensic body fluid identification
Application notes
TaqMan® Array Cards for combined mRNA and miRNA analysis
Endogenous controls for real-time quantitation of miRNA using TaqMan® MicroRNA assays
High correlation of miRNA quantitation data from matched FFPE and snap-frozen tissues using TaqMan® MicroRNA Assays
Optimized protocol with low sample input for profiling human microRNA using the OpenArray® platform
Optimized blood plasma protocol for profiling human miRNAs using the OpenArray® Real-Time PCR System
Posters
MicroRNA quantitation by real-time PCR
TaqMan®-based miRNA profiles classify embryonic stem and differentiated cells
Custom RT and PreAmp pools using TaqMan® miRNA Assays
Ready to order TaqMan® MicroRNA Assays?
Ordering TaqMan® MicroRNA Assays is quick and easy with the TaqMan® Assay search tool. Choose from mature miRNA assays, pre-miRNA assays for over 200 species, as well as mimics, inhibitors, and controls. Access the assay search tool to order TaqMan® MicroRNA Assays, or watch a video to learn how to search and order TaqMan® MicroRNA Assays.
For Research Use Only. Not for use in diagnostic procedures.